MIRIAM URIs
An important role of the MIRIAM guidelines consists of their use in the controlled annotation of model components, based on Uniform Resource Identifiers (URIs). In support of this task, a set of controlled URIs were created: MIRIAM URIs. These allow the unique and unambiguous identification of a component in a stable and perennial manner. The #MIRIAM Registry and Identifiers.org system are a set of services and resources that provide support for generating, interpreting and resolving MIRIAM URIs.
MIRIAM URIs are composed of two main parts: the first defines a namespace that particular 'entities of the same type' may occupy. This is called a 'collection'. The second part precisely identifies a given entity within this collection, and is called a 'record'. For example, 'http://identifiers.org/pubmed/16333295' is the MIRIAM URI that identifies the publication of the MIRIAM Standard within the PubMed data collection. Here, 'http://identifiers.org/pubmed' defines the collection (PubMed), and '16333295' precisely identifies the entity record within it.
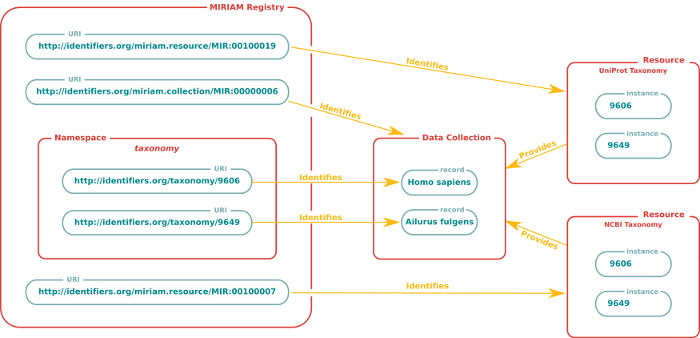
The scope or domain of each data collection is strictly defined, and where a resource references many types of data (for example genes and proteins), the scope is clearly demarcated, and is reflected in URIs when necessary, for instance by 'sub-classing' the collections provided by the resource. For example, the Kyoto Encyclopedia of Genes and Genomes, is been divided into different collections, all of them being part of the kegg "class" (which can be seen in the URI):
An example of the use of MIRIAM URIs in an RDF annotation is provided below:

The example above describes a complex (usage of the bqbiol:hasPart qualifier), composed of:
- P69905 (usage of the MIRIAM URI http://identifiers.org/uniprot/P69905)
- P68871 (usage of the MIRIAM URI http://identifiers.org/uniprot/P68871)
- CHEBI:17627 (usage of the MIRIAM URI http://identifiers.org/obo.chebi/CHEBI:3A17627)
The annotation is linked to the proper model element by using a unique identifier. In order to actually use the piece of annotation above, one would need to replace heme, with the unique identifier of the model element defining the complex.
More detailed examples showing how to use this service to identify collections, resources and records are described here.
Indirectly Resolvable MIRIAM URNs
The original MIRIAM URIs were no resolvable directly in a web browser, but required the use of web services. As with the identifiers.org URL scheme, this URN system is also based upon the information stored in the MIRIAM Registry, with the resolving framework being provided by identifiers.org. For example, the 'heme' complex annotation illustrated above could equally be written (mouseover to view the URN identifier form):
- P69905 (using indirectly resolvable URI urn:miriam:uniprot:P69905)
- P68871 (using indirectly resolvable URI urn:miriam:uniprot:P68871)
- CHEBI:17627 (using indirectly resolvable URI urn:miriam:obo.chebi:CHEBI%3A17627)
Note that the identifiers used by many data providers tend to be of a form where there is a prefix identifying the database, followed by a separator, and ending with some alphanumeric key to uniquely identify a record. The separator in many cases is either an 'underscore', or a 'colon'. In the case of MIRIAM URNs, the colon character is used as a delimiter between the different parts of the URN and therefore is considered a "reserved" character. In accordance to the URI Generic Syntax, this character must be percent-encoded. Therefore, any ':' in the identifier part of a MIRIAM URN must always be encoded as '%3A'.
Although the URL form is now strongly advocated, both URN and URL forms are supported equally.
MIRIAM Registry and Identifiers.org
In order to enable interoperability of this annotation scheme, the community has to agree upon a set of recognised collections. The MIRIAM Registry is an online service created to catalogue these collections, their URIs and the corresponding physical URLs or resources, whether they are controlled vocabularies or databases.
By using the MIRIAM Registry, one can (via Web Services) generate MIRIAM URIs (URN form), as well as resolve them (transform them into physical locations of the corresponding pieces of knowledge). Directly resolvable URIs, these are also available through Identifiers.org, which acts as a resolving layer above the the Registry. Both forms will be supported equally, with services provided to allow interconversion between them.
Publications
- Juty N., Le Novère N., Laibe C. (2012)
- Identifiers.org and MIRIAM Registry: community resources to provide persistent identification.
- Nucleic Acids Research, 40: D580-D586
- PubMedOpen Access
- Laibe C., Le Novère N. (2007)
- MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology.
- BMC Systems Biology, 1: 58
- PubMed - Open Access
- Le Novère N, Courtot M, Laibe C (2007)
- Adding semantics in kinetics models of biochemical pathways.
- Proceedings of the 2nd International Symposium on experimental standard conditions of enzyme characterizations, available at http://www.beilstein-institut.de/index.php?id=196
